Isolation, selection and characterisation of microorganisms with potential to form unique consortium for wine production
DOI:
https://doi.org/10.18832/kp2024.70.855Keywords:
isolation of microorganisms, characterisation of microorganisms, yeasts, lactic acid bacteriaAbstract
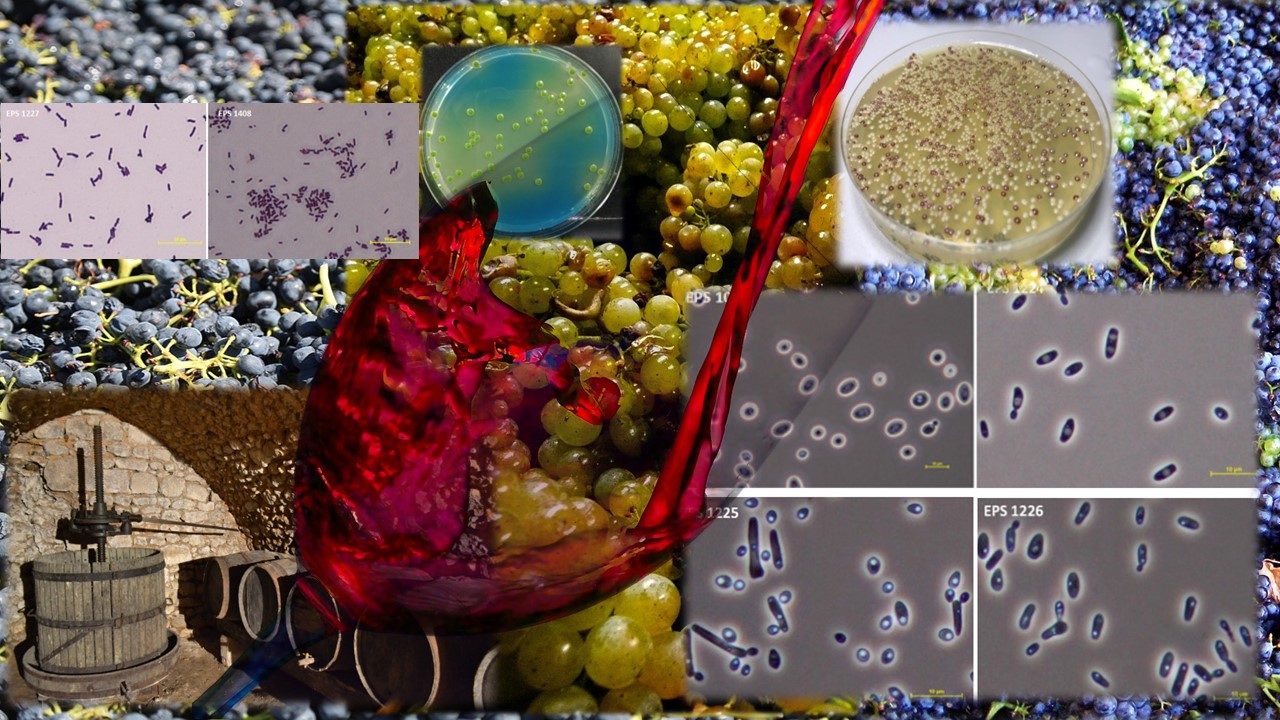
This paper focuses on a selection of microorganisms that should eventually form a Defined Consortium of Wine Microorganisms. The consortium might serve as a sophisticated oenological product for the production of wine with attractive organoleptic features.
The Defined Consortium of Wine Microorganisms was obtained from non-saccharomyces and saccharomyces yeasts and lactic acid bacteria isolated from places where autochthonous microflora can be expected to occur (grape berries, wine lees or cellar spaces). A total of 42 microorganisms were obtained by surface smears, fallouts from the air or isolation from fermented must. All isolates were then tested and the best strains were selected on the basis of technological and phenotypic characterization using standard microbiological techniques. The main criteria for the selection of strains were the ability to ferment and the production of organoleptically active compounds. Sulphur dioxide or ethanol tolerance, β-glucosidase activity, a taxonomic identification, the tendency to produce sulphane or the ability of lactic acid bacteria to perform malolactic fermentation were also considered. According to these results, the non-saccharomyces yeast Starmerella bacillaris, the saccharomyces yeast Saccharomyces cerevisiae and the lactic acid bacterium Levilactobacillus brevis were selected as part of the Defined Consortium of Wine Microorganisms.
References
Aranda, A., Matallana, E., del Olmo, M. (2011). Saccharomyces Yeasts I: Primary Fermentation. In: Carrascosa, A.V., Muñoz, R., González, R. (eds.) Molecular wine microbiology, Elsevier, Cham, Switzerland, p. 2. https://doi.org/10.1016/B978-0-12-375021-1.10001-3
Box, G.E.P., Cox, D.R. (1964). An analysis of transformations. Journal of the Royal Statistical Society: Series B, 26(2), 211–243. https://doi.org/10.1111/j.2517-6161.1964.tb00553.x
Camilo, S., Chandra, M., Branco, P., Malfeito-Ferreira, M. (2022). Wine Microbial Consortium: Seasonal Sources and Vectors Linking Vineyard and Winery Environments. Fermentation, 8(7), 324. https://doi.org/10.3390/fermentation8070324
Castellucci, F. (2012). Guidelines for the Characterization of Wine Yeasts of the Genus Saccharomyces Isolated from Vitivinicultural Environments. International Organisation of Vine and Wine: Paris, France, p.21. Available from: https://www.oiv.int/public/medias/1429/oivoeno-370-2012-en.pdf
Cavaglia, J., Garcia, S.M., Roger, J.-M., Mestres, M., Boqué, R. (2022). Detection of bacterial spoilage during wine alcoholic fermentation using ATR-MIR and MCR-ALS. Food Control, 142, 109269. https://doi.org/10.1016/j.foodcont.2022.109269
Ciani, M., Comitini, F., Mannazzu, I., Domizio, P. (2010). Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Research, 10(2), 123–133. https://doi.org/10.1111/j.1567-1364.2009.00579.x
Costello, P.J., Déléris-Bou, M., Descenzo, R., Hall, N., Krieger, S., Lonvaud-Funel, A., Loubser, P., Heras, J.M. et al. (2015). Malolactic Fermentation - Importance of Wine Lactic Acid Bacteria in Winemaking. Lallemand Incorporated, 159–164. ISBN 978-2-9815255-0-5; available from: https://www.lallemandwine.com/wp-content/uploads/2015/10/Lallemand-Malolactic-Fermentation.pdf
Dalgaard, P. (Producer). (2010). R Development Core Team (2010): R: A language and environment for statistical computing. Computer programme http://www.R-project.org/
Englezos, V., Giacosa, S., Rantsiou, K., Rolle, L., Cocolin, L. (2017). Starmerella bacillaris in winemaking: Opportunities and risks. Current Opinion in Food Science, 17, 30–35. https://doi.org/10.1016/j.cofs.2017.08.007
Fia, G. (2016). Wine lees: traditional and potential innovative techniques for their exploitation in winemaking. In: Morata, A., Loira, I. (eds.) Grape and wine biotechnology, InTech Open, 345–359. https://doi.org/10.5772/65043
Gomes, R.J., de Fatima Borges, M., de Freitas Rosa, M., Castro-Goméz, R.J.H., Spinosa, W.A. (2018). Acetic acid bacteria in the food industry: systematics, characteristics and applications. Food technology and biotechnology, 56(2), 139–151. https://doi.org/10.17113/ftb.56.02.18.5593
Kalhotka, L., Dostálová, L., Detvanová, L. (2015). Potravinářská mikrobiologie – návody do cvičení, p. 44. ISBN 978-80-7509-259-5; available in Czech from: https://web2.mendelu.cz/af_291_projekty/files/23/23-potravinarska_mikrobiologie__navody_do_cviceni-pmzii.pdf
Kántor, A., Mareček, J., Ivanišová, E., Terentjeva, M., Kačániová, M. (2017). Microorganisms of grape berries. Proceedings of the Latvian academy of sciences, 71(6), 502–508. https://doi.org/10.1515/prolas-2017-0087
Li, H., Jiang, D., Liu, W., Yang, Y., Zhang, Y., Jin, C., Sun, S. (2020). Comparison of fermentation behaviors and properties of raspberry wines by spontaneous and controlled alcoholic fermentations. Food Research International, 128, 108801. https://doi.org/10.1016/j.foodres.2019.108801
Nilsson, R.H., Larsson, K.-H., Taylor, A.F.S., Bengtsson-Palme, J., Jeppesen, T.S., Schigel, D., Kennedy, P., Picard, K. et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research, 47(D1), D259–D264. https://doi.org/10.1093/nar/gky1022
OIV Codex (2018). International Code of Oenological Practices. Resolution OIV-Oeno 591A-2018. Free sulphur dioxide – update to method OIVMA-AS323-04A. Available from: https://www.oiv.int/node/3139/download/pdf
OIV Codex (2021). International Code of Oenological Practices, Issue 2021, pp. 435. ISBN 978-2-85038-030-3; available from: https://www.oiv.int/public/medias/7713/en-oiv-code-2021.pdf
Říhová Ambrožová J., Trögl J. (2014). Standardní postupy environmentální mikrobiologie. Návody úloh a laboratorních cvičení, p.78. ISBN 978-80-7414-854-5 (online); avalible in Czech from: http://envimod.fzp.ujep.cz/sites/default/files/skripta/33e_final_tisk.pdf
Ritz, C., Strebig, J.C. (2016). Analysis of dose-response curves. R package version, 3.0-1.
Strahsburger, E., de Lacey, A.M.L., Marotti, I., DiGioia, D., Biavati, B., Dinelli, G. (2017). In vivo assay to identify bacteria with β-glucosidase activity. Electronic Journal of Biotechnology, 30, 83–87. https://doi.org/10.1016/j.ejbt.2017.08.010
Yoon, S.-H., Ha, S.-M., Kwon, S., Lim, J., Kim, Y., Seo, H., Chun, J. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. International Journal of Systematic and Evolutionary Microbiology, 67(5), 1613–1617. https://doi.org/10.1099/ijsem.0.001755
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Adrienn Dallos, Jana Juříková, Vít Paulíček, Martin Gajdošík, Vendula Pavelková, Veronika Andrýsková

This work is licensed under a Creative Commons Attribution 4.0 International License.







